See the Nephele User Guide for how to submit a job. Below are the particular choices we made at each step outlined in the guide for the workshop workflow.
QC job
We recommend running the QC pipeline with just default settings for any dataset you will analyze. This will give you a FastQC report with information about the quality of the data. Then you can proceed to running one of the analysis pipelines (below).
- Step 1 - From the Front Page of Nephele, click on tab “1. Pre-Process”
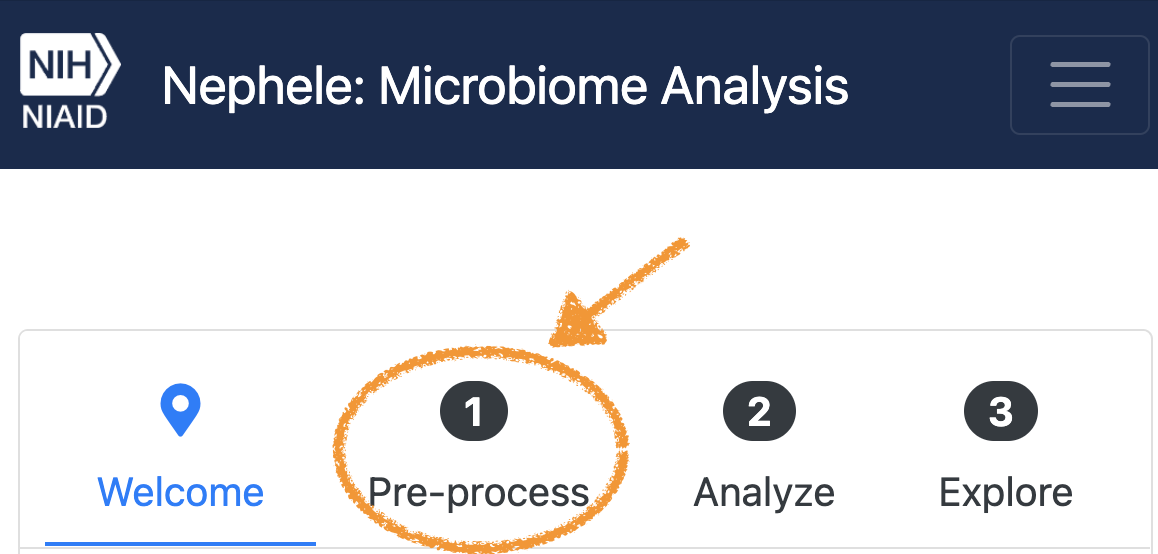 . Then click “Start Job.”
. Then click “Start Job.” - Step 2 - Choose Paired End FASTQ.
- Step 3 - Click on Upload via Globus (the workshop data is stored on a NIAID server, and we will be transferring to Nephele using the Globus service which NIH subscribes to, and which many other institutions do as well). From the Materials page, click on the Globus link for the dataset you want to analyze. It will be copied to your clipboard. Go back to the Nephele Globus upload page, and paste in the link.
- Step 4 - On the workshop Materials page, click on the mapping file link for your dataset, it will download to your computer. Back on the Nephele page, upload this file as it contains the metadata for your job. Nephele will check and see if the files listed in the mapping file are in the Globus directory from the link before proceeding.
- Step 5 - Click Select for the QC pipeline.
- Step 6 - Put in a name for your job and click “Validate and Submit”. We will be running with default parameters, but you can also click on the “Options tab” and input additional trimming or merging options).
- You should get an email for when the job starts and another when it ends. The results will have the MultiQC report we looked at in the workshop.
16S job
- Step 1 - From the Front Page of Nephele, click on tab “2. Analyze tab”
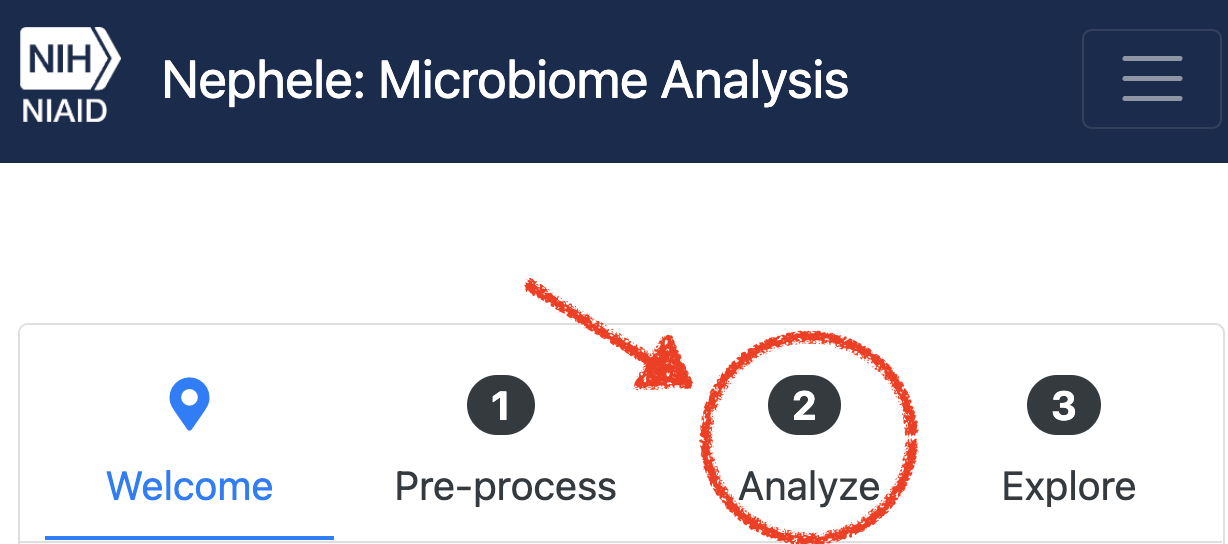 . Then in the first column, under “Amplicon metagenomics”, choose “Start Job” for the DADA2 pipeline.
. Then in the first column, under “Amplicon metagenomics”, choose “Start Job” for the DADA2 pipeline. - Follow the same procedure as we did above for Steps 2-5 in the QC job to choose the paired end data type, upload via Globus, upload the 16S mapping file, and select the pipeline.
- Step 6 - For the pipeline parameters, we put in a name for the job. On the “Pre-processing” tab under “Filter and Trim,” fill in the Trim left forward and Trim left reverse trims with 10. This is the number of base pairs we will trim from the beginning of each read. Under “Merge Pairs,” check the box for Trim overhanging sequence.
- Under the “Analysis” tab, choose the SILVA v138.1 database (or SILVA v132 which is the older version used in the results of the workshop, though we recommend the newer version going forward). Then click “Validate and Submit”
WGS job
- Step 1 - From the Front Page of Nephele, click on tab “2. Analyze tab”
 . Then in the second column, under “Shotgun metagenomics”, choose “Start Job” for the WGSA2 pipeline.
. Then in the second column, under “Shotgun metagenomics”, choose “Start Job” for the WGSA2 pipeline. - Follow the same procedure as we did above for Steps 2-5 in the QC job to choose the paired end data type, upload via Globus (using the Globus link for the WGS HMP dataset), upload the WGS mapping file, and select the pipeline.
- Step 6 - for the pipeline parameters, give the job a name, check Run AMR Finder, choose MetaCyc database for Metabolic pathways Database, and click “Validate and Submit.”



